Cluster and Outlier Analysis, Grouping Analysis and Hot Spot Analysis Tools
Cluster and Outlier Analysis (Anselin Local Morans I)
How to use Cluster and Outlier Analysis (Anselin Local Morans I) Tool in Arc Toolbox??
.webp) |
| Cluster and Outlier Analysis (Anselin Local Morans I) |
Path to access the tool
:
Cluster
and Outlier Analysis (Anselin Local Morans I) Tool, Mapping Clusters Toolset,
Spatial Statistics Tools Toolbox
Cluster and Outlier Analysis (Anselin
Local Morans I)
Given a set of weighted
features, identifies statistically significant hot spots, cold spots, and
spatial outliers using the Anselin Local Moran's I statistic.
1. Input Feature Class
The feature class for
which cluster and outlier analysis will be performed.
2. Input Field
The numeric field to be
evaluated.
3. Output Feature Class
The output feature class
to receive the results fields.
4. Conceptualization of Spatial Relationships
Specifies how spatial relationships among features are defined.
- INVERSE_DISTANCE—Nearby neighboring features have a larger influence on the computations for a target feature than features that are far away.
- INVERSE_DISTANCE_SQUARED—Same asINVERSE_DISTANCE except that the slope is sharper, so influence drops off more quickly, and only a target feature's closest neighbors will exert substantial influence on computations for that feature.
- FIXED_DISTANCE_BAND—Each feature is analyzed within the context of neighboring features. Neighboring features inside the specified critical distance (Distance Band or Threshold Distance) receive a weight of one and exert influence on computations for the target feature. Neighboring features outside the critical distance receive a weight of zero and have no influence on a target feature's computations.
- ZONE_OF_INDIFFERENCE—Features within the specified critical distance (Distance Band or Threshold Distance) of a target feature receive a weight of one and influence computations for that feature. Once the critical distance is exceeded, weights (and the influence a neighboring feature has on target feature computations) diminish with distance.
- CONTIGUITY_EDGES_ONLY—Only neighboring polygon features that share a boundary or overlap will influence computations for the target polygon feature.
- CONTIGUITY_EDGES_CORNERS—Polygon features that share a boundary, share a node, or overlap will influence computations for the target polygon feature.
- GET_SPATIAL_WEIGHTS_FROM_FILE—Spatial relationships are defined by a specified spatial weights file. The path to the spatial weights file is specified by the Weights Matrix File parameter.
5. Distance Method
Specifies how distances are calculated from each feature to neighboring features.
- EUCLIDEAN_DISTANCE—The straight-line distance between two points (as the crow flies)
- MANHATTAN_DISTANCE—The distance between two points measured along axes at right angles (city block); calculated by summing the (absolute) difference between the x- and y-coordinates
6. Standardization
Row standardization is recommended whenever the distribution of your features is potentially biased due to sampling design or an imposed aggregation scheme.
- NONE—No standardization of spatial weights is applied.
- ROW—Spatial weights are standardized; each weight is divided by its row sum (the sum of the weights of all neighboring features).
7. Distance Band or Threshold Distance (optional)
Specifies a cutoff
distance for Inverse Distance and Fixed Distance options. Features outside the
specified cutoff for a target feature are ignored in analyses for that feature.
However, for Zone of Indifference, the influence of features outside the given
distance is reduced with distance, while those inside the distance threshold
are equally considered. The distance value entered should match that of the
output coordinate system.
For the Inverse Distance
conceptualizations of spatial relationships, a value of 0 indicates that no
threshold distance is applied; when this parameter is left blank, a default
threshold value is computed and applied.
This default value is the Euclidean
distance that ensures every feature has at least one neighbor.
This parameter has no
effect when Polygon Contiguity or Get Spatial Weights From File spatial
conceptualizations are selected.
8. Weights Matrix File (optional)
The path to a file
containing weights that define spatial, and potentially temporal, relationships
among features.
9. Apply False Discovery Rate (FDR) Correction (optional)
Specifies whether statistical significance will be assessed with or without FDR correction.
- Checked—Statistical significance will be based on the False Discovery Rate correction for a 95 percent confidence level.
- Unchecked—Features with p-values less than 0.05 will appear in the COType field reflecting statistically significant clusters or outliers at a 95 percent confidence level. This is the default.
10. Number of Permutations (optional)
The number of random permutations for the calculation of pseudo p-values. The default number of permutations is 499. If you choose 0 permutations, the standard p-value is calculated.
- 0—Permutations are not used and a standard p-value is calculated.
- 99—With 99 permutations, the smallest possible pseudo p-value is 0.01 and all other pseudo p-values will be even multiples of this value.
- 199—With 199 permutations, the smallest possible pseudo p-value is 0.005 and all other possible pseudo p-values will be even multiples of this value.
- 499—With 499 permutations, the smallest possible pseudo p-value is 0.002 and all other pseudo p-values will be even multiples of this value.
- 999—With 999 permutations, the smallest possible pseudo p-value is 0.001 and all other pseudo p-values will be even multiples of this value.
- 9999—With 9999 permutations, the smallest possible pseudo p-value is 0.0001 and all other pseudo p-values will be even multiples of this value.
Grouping Analysis
How to use Grouping Analysis Tool in Arc Toolbox??
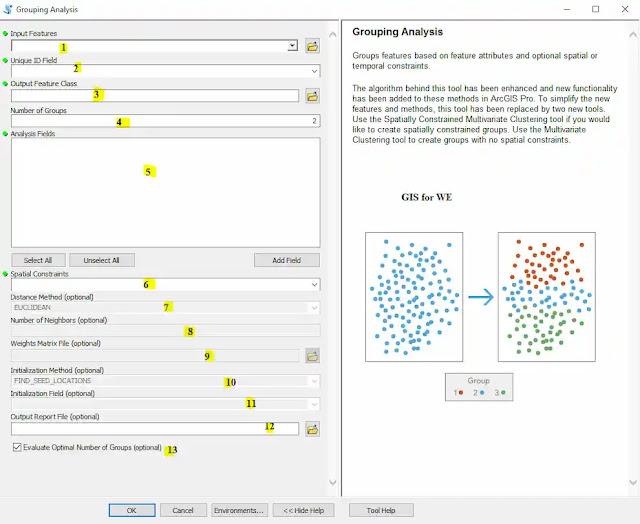 |
| Grouping Analysis |
Path to access the tool
:
Grouping
Analysis Tool, Mapping Clusters Toolset, Spatial Statistics Tools
Toolbox
Grouping Analysis
Groups features based on
feature attributes and optional spatial or temporal constraints.
The algorithm behind
this tool has been enhanced and new functionality has been added to these
methods in ArcGIS Pro. To simplify the new features and methods, this tool has
been replaced by two new tools. Use the Spatially Constrained Multivariate
Clustering tool if you would like to create spatially constrained groups. Use
the Multivariate Clustering tool to create groups with no spatial constraints.
1. Input Features
The feature class or
feature layer for which you want to create groups.
2. Unique ID Field
An integer field
containing a different value for every feature in the input feature class. If
you don't have a Unique ID field, you can create one by adding an integer field
to your feature class table and calculating the field values to equal the FID
or OBJECTID field.
3. Output Feature Class
The new output feature
class created containing all features, the analysis fields specified, and a
field indicating to which group each feature belongs.
4. Number of Groups
The number of groups to
create. The Output Report parameter will be disabled for more than 15 groups.
5. Analysis Fields
A list of fields you
want to use to distinguish one group from another. The Output Report parameter
will be disabled for more than 15 fields.
6. Spatial Constraints
Specifies if and how spatial relationships among features should constrain the groups created.
- CONTIGUITY_EDGES_ONLY—Groups contain contiguous polygon features. Only polygons that share an edge can be part of the same group.
- CONTIGUITY_EDGES_CORNERS—Groups contain contiguous polygon features. Only polygons that share an edge or a vertex can be part of the same group.
- DELAUNAY_TRIANGULATION—Features in the same group will have at least one natural neighbor in common with another feature in the group. Natural neighbor relationships are based on Delaunay Triangulation. Conceptually, Delaunay Triangulation creates a nonoverlapping mesh of triangles from feature centroids. Each feature is a triangle node and nodes that share edges are considered neighbors.
- K_NEAREST_NEIGHBORS—Features in the same group will be near each other; each feature will be a neighbor of at least one other feature in the group. Neighbor relationships are based on the nearest K features, where you specify an Integer value, K, for the Number of Neighbors parameter.
- GET_SPATIAL_WEIGHTS_FROM_FILE—Spatial, and optionally temporal, relationships are defined by a spatial weights file (.swm). Create the spatial weights matrix file using the Generate Spatial Weights Matrix tool or the Generate Network Spatial Weights tool.
- NO_SPATIAL_CONSTRAINT—Features will be grouped using data space proximity only. Features do not have to be near each other in space or time to be part of the same group.
7. Distance Method (optional)
Specifies how distances are calculated from each feature to neighboring features.
- EUCLIDEAN—The straight-line distance between two points (as the crow flies)
- MANHATTAN—The distance between two points measured along axes at right angles (city block); calculated by summing the (absolute) difference between the x- and y-coordinates
8. Number of Neighbors (optional)
This parameter is
enabled whenever the Spatial Constraints parameter is K_NEAREST_NEIGHBORS or
one of the contiguity methods (CONTIGUITY_EDGES_ONLY or
CONTIGUITY_EDGES_CORNERS). The default number of neighbors is 8 and cannot be
smaller than 2 for K_NEAREST_NEIGHBORS. This value reflects the exact number of
nearest neighbor candidates to consider when building groups. A feature will
not be included in a group unless one of the other features in that group is a
K nearest neighbor. The default for CONTIGUITY_EDGES_ONLY and
CONTIGUITY_EDGES_CORNERS is 0. For the contiguity methods, this value reflects
the minimum number of neighbor candidates to consider. Additional nearby
neighbors for features with less than the Number of Neighbors specified will be
based on feature centroid proximity.
9. Weights Matrix File (optional)
The path to a file
containing spatial weights that define spatial relationships among features.
10. Initialization Method (optional)
Specifies how initial seeds are obtained when the Spatial Constraint parameter selected is NO_SPATIAL_CONSTRAINT. Seeds are used to grow groups. If you indicate you want three groups, for example, the analysis will begin with three seeds.
- FIND_SEED_LOCATIONS—Seed features will be selected to optimize performance.
- GET_SEEDS_FROM_FIELD—Nonzero entries in the Initialization Field will be used as starting points to grow groups.
- USE_RANDOM_SEEDS—Initial seed features will be randomly selected.
11. Initialization Field (optional)
The numeric field
identifying seed features. Features with a value of 1 for this field will be
used to grow groups.
12. Output Report File (optional)
The full path for the
PDF report file to be created summarizing group characteristics. This report
provides a number of graphs to help you compare the characteristics of each
group. Creating the report file can add substantial processing time.
13. Evaluate Optimal Number of Groups (optional)
Specifies whether the tool will assess the optimal number of groups, 2 through 15.
- Checked—Groupings from 2 to 15 will be evaluated.
- Unchecked—No evaluation of the number of groups will be performed. This is the default.
Hot Spot Analysis (Getis-Ord Gi*)
How to use Hot Spot Analysis (Getis-Ord Gi*) Tool in Arc Toolbox??
.webp) |
| Hot Spot Analysis (Getis-Ord Gi*) |
Path to access the tool
:
Hot
Spot Analysis (Getis-Ord Gi*) Tool, Mapping Clusters Toolset, Spatial
Statistics Tools Toolbox
Hot Spot Analysis (Getis-Ord Gi*)
Given a set of weighted
features, identifies statistically significant hot spots and cold spots using
the Getis-Ord Gi* statistic.
1. Input Feature Class
The feature class for
which hot spot analysis will be performed.
2. Input Field
The numeric field
(number of victims, crime rate, test scores, and so on) to be evaluated.
3. Output Feature Class
The output feature class
to receive the z-score and p-value results.
4. Conceptualization of Spatial Relationships
Specifies how spatial relationships among features are defined.
- INVERSE_DISTANCE—Nearby neighboring features have a larger influence on the computations for a target feature than features that are far away.
- INVERSE_DISTANCE_SQUARED—Same asINVERSE_DISTANCE except that the slope is sharper, so influence drops off more quickly, and only a target feature's closest neighbors will exert substantial influence on computations for that feature.
- FIXED_DISTANCE_BAND—Each feature is analyzed within the context of neighboring features. Neighboring features inside the specified critical distance (Distance Band or Threshold Distance) receive a weight of one and exert influence on computations for the target feature. Neighboring features outside the critical distance receive a weight of zero and have no influence on a target feature's computations.
- ZONE_OF_INDIFFERENCE—Features within the specified critical distance (Distance Band or Threshold Distance) of a target feature receive a weight of one and influence computations for that feature. Once the critical distance is exceeded, weights (and the influence a neighboring feature has on target feature computations) diminish with distance.
- CONTIGUITY_EDGES_ONLY—Only neighboring polygon features that share a boundary or overlap will influence computations for the target polygon feature.
- CONTIGUITY_EDGES_CORNERS—Polygon features that share a boundary, share a node, or overlap will influence computations for the target polygon feature.
- GET_SPATIAL_WEIGHTS_FROM_FILE—Spatial relationships are defined by a specified spatial weights file. The path to the spatial weights file is specified by the Weights Matrix File parameter.
5. Distance Method
Specifies how distances are calculated from each feature to neighboring features.
- EUCLIDEAN_DISTANCE—The straight-line distance between two points (as the crow flies)
- MANHATTAN_DISTANCE—The distance between two points measured along axes at right angles (city block); calculated by summing the (absolute) difference between the x- and y-coordinates
6. Standardization
Row standardization has no impact on this tool: results from Hot Spot Analysis (the Getis-Ord Gi* statistic) would be identical with or without row standardization. The parameter is disabled; it remains as a tool parameter only to support backwards compatibility.
- NONE—No standardization of spatial weights is applied.
- ROW—No standardization of spatial weights is applied.
7. Distance Band or Threshold Distance (optional)
Specifies a cutoff
distance for Inverse Distance and Fixed Distance options. Features outside the
specified cutoff for a target feature are ignored in analyses for that feature.
However, for Zone of Indifference, the influence of features outside the given
distance is reduced with distance, while those inside the distance threshold
are equally considered. The distance value entered should match that of the
output coordinate system.
For the inverse distance
conceptualizations of spatial relationships, a value of 0 indicates that no
threshold distance is applied; when this parameter is left blank, a default
threshold value is computed and applied. This default value is the Euclidean
distance that ensures every feature has at least one neighbor.
This parameter has no
effect when polygon contiguity (CONTIGUITY_EDGES_ONLY or
CONTIGUITY_EDGES_CORNERS) or GET_SPATIAL_WEIGHTS_FROM_FILE spatial
conceptualizations are selected.
8. Self Potential Field (optional)
The field representing
self potential: the distance or weight between a feature and itself.
9. Weights Matrix File (optional)
The path to a file
containing weights that define spatial, and potentially temporal, relationships
among features.
10. Apply False Discovery Rate (FDR) Correction (optional)
Specifies whether statistical significance will be assessed with or without FDR correction.
- Checked—Statistical significance will be based on the False Discovery Rate correction.
- Unchecked—Statistical significance will be based on the p-value and z-score fields. This is the default.

Comments
Post a Comment